Research Activity
When citing a core facility, please ensure that you are referrencing their Research Resource Identifier (RRID) number, and funding source. A citation example for the Cryo-EM facility is provided below:
"Funding to establish the Cryo-EM Core Facility (RRID:SCR_026324) was provided by the National Science Foundation, the Murdock Charitable Trust, and MSU's Office of Research and Economic Development, and Graduate Education"
If users work closely with a member of facility staff on a project, they are expected to be viewed as collaborators. Please include them in attributions, and in relevant project meetings.
Structures Determined by Facility Users
|
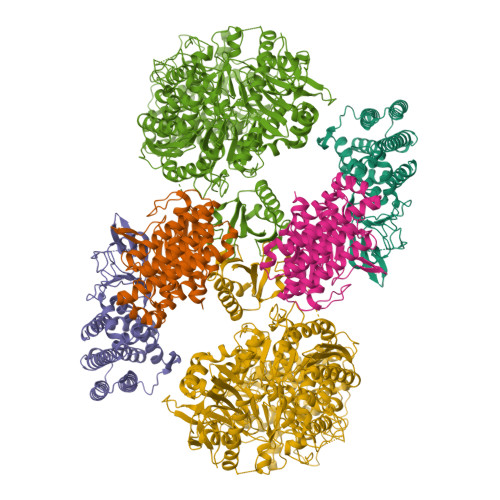
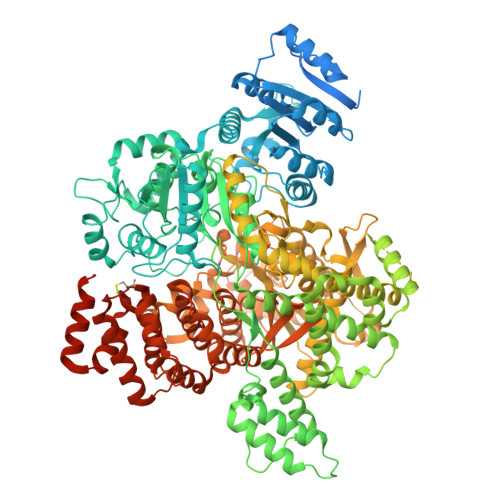
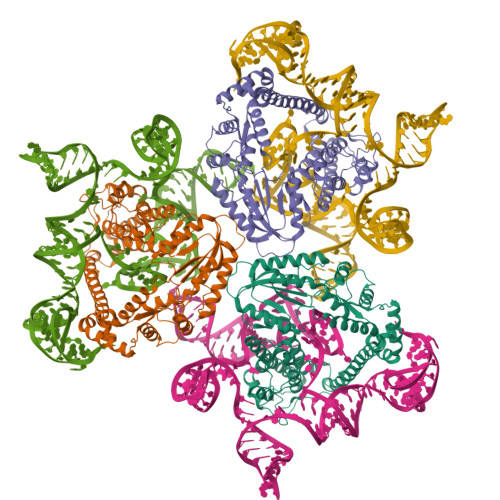
Cas1-Cas2/3 integrase, heterohexameric assemblyPDB ID: 9P11 Resolution: 3.50 Å |
Cas1-Cas2/3 integrase bound to foreign dsDNA fragmentPDB ID: 9P1D Resolution: 3.31 Å |
|
Structure of thioferritin exhibiting iron mineral nucleation, from Pyrococcus furiosisPDB ID: 9CZ8 Resolution: 1.91 Å |
Structure of a widespread heme dechelatase in healthy & pathogenic human microbiomesPDB ID: 9D26 Resolution: 2.60 Å |
|
HmuS heme dechelatase: disordered domain 1, heme freePDB ID: 9P4S Resolution: 2.55 Å |
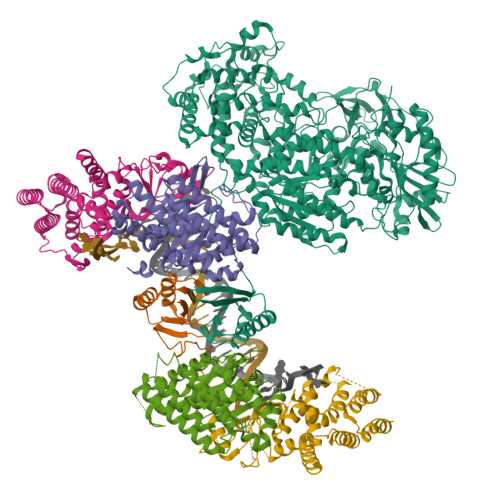
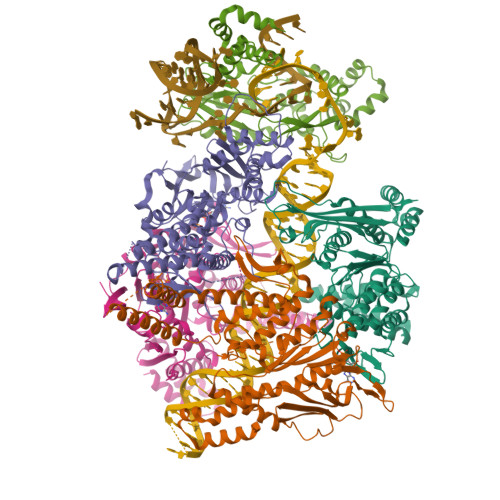
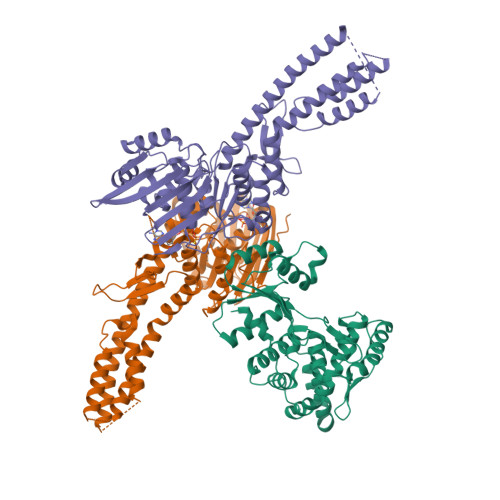
Structure of the Retron IA Complex without the HNH NucleasePDB ID: 9N6C Resolution: 2.99 Å |
|
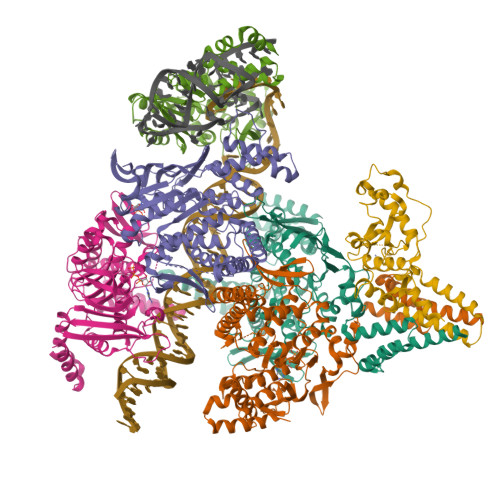
Structure of the retron IA complex with HNH nuclease in the "up" orientationPDB ID: 9N6B Resolution: 3.09 Å |
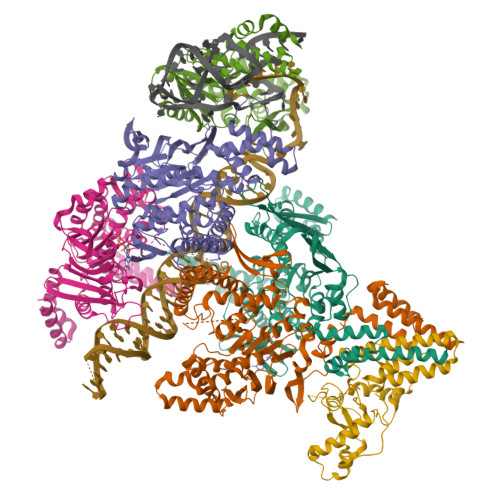
Structure of the retron IA complex with HNH nuclease in the "down" orientationPDB ID: 9N69 Resolution: 3.13 Å |
|
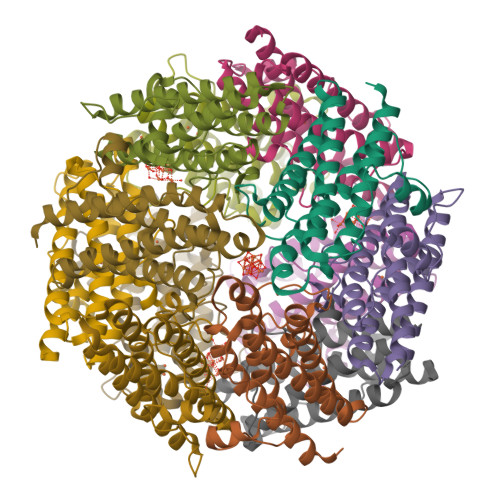
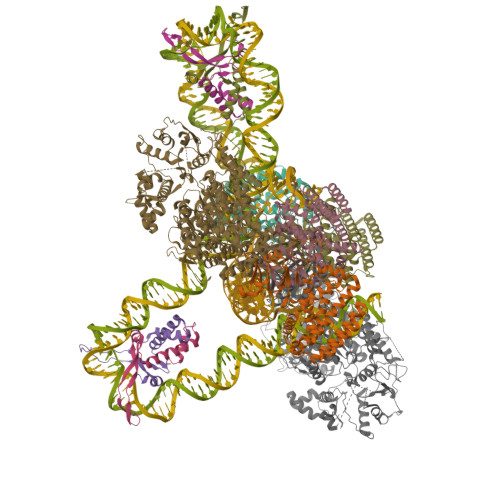
Cryo-EM structure of the trimeric SenDRT9 RT-ncRNAPDB ID: 9NLX Resolution: 3.20 Å |
Cryo-EM structure of hexameric SenDRT9 RT-ncRNA complexPDB ID: 9NLV Resolution: 2.60 Å |
|
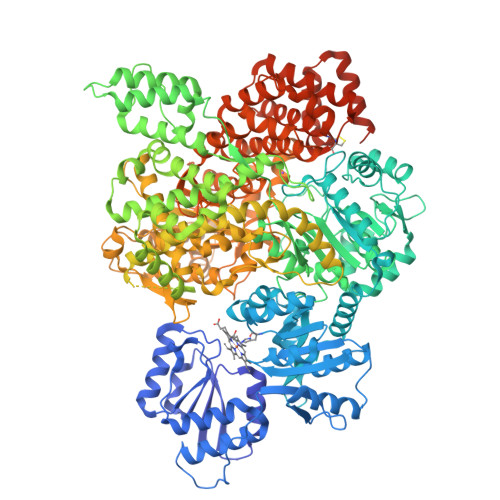
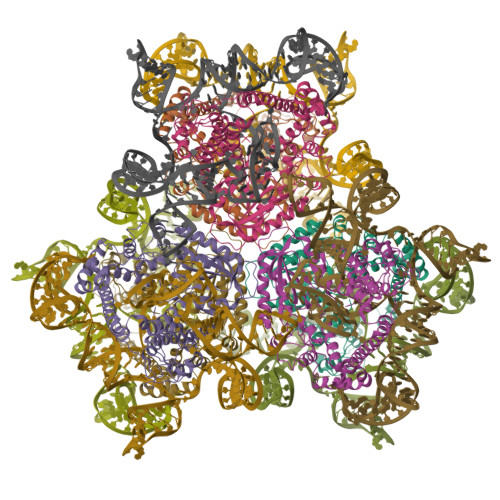
Asymmetric unit of the PARIS Immune Complex at 3.2 Angstrom ResolutionPDB ID: 8UX9 Resolution: 3.20 Å |
Cas1-Cas2/3 integrase and IHF bound to CRISPR leader, repeat and foreign DNAPDB ID: 8FLJ Resolution: 3.48 Å |
2025 Publications
George, J.T., Burman, N., Wilkinson, R.A. et al. Structural basis of antiphage defence by an ATPase-associated reverse transcriptase. Nat Commun 16, 8459 (2025). https://doi.org/10.1038/s41467-025-63285-6